… I came to see the owls as one of countless shapes the forest assumes, more than as an animal that resides in a forest. In this view, if the organism is removed from the old growth, it ceases to be a spotted owl and becomes just a brown, speckled, dark-eyed, meat-eating bird.
The feature that most exemplifies this awareness for me is the owl’s feathers. Unlike most raptors in this region, northern spotted owls don’t migrate south in the winter. They are here in rain and snow and cold. Yet they are not exceptional thermoregulators as one would expect of a nonmigrant facing a rainy, snowy Cascade Mountain winter.
The reason for this counterintuitive disparity is the old-growth forest, which buffers temperature extremes at both ends of the spectrum by as much as twenty degrees Fahrenheit in relation to adjacent clearings. The owls wear old growth like another layer of feathers. That is, their preferred habitat provides sufficient thermal protection, so that they do not have to expend energy growing as much down as they would need if they dwelled in open country.
The deep multilayered canopy characteristic of old-growth groves also intercepts enough snow to permit the owl’s preferred prey species – the northern flying squirrel – to remain active all winter long, feeding on truffle mushrooms on the open ground at the bottom of snow wells around the bases of the trees. With food and warmth (as well as many other life needs) literally covered by the old growth, the owl does not have to make a long flight to warmer climes at the onset of autumn.
Instead, the owl makes short flights in response to immediate conditions. Sun gaps on otherwise snowy January days draw them out from beneath sheltering midcanopy mistletoe umbrellas to ascend to high branches, where direct solar radiance can offer springlike warmth even during the coldest time of year. And in the heat of August, the owls are often found perched low in vine maples a few feet above a cooling creek in shady northeast- facing drainages. This behavioural thermoregulation and the incorporation of the forest itself into their meaningful physiology provide just two of many possible examples that demonstrate why efforts to reduce the spotted owl to a bird in a habitat represents extreme oversimplification.
from The Mountain Lion by Tim Fox, excerpted in Forest Under Story
On the other hand
Internet home of C. Susannah Tysor
Author Archives: susannah
Keeping up with the literature (in your own lab)
My lab has started blogging about each other’s papers. People in our lab use quite a variety of techniques and have some pretty different research focuses. I love seeing my labmate’s work come together into a cohesive whole and I love digging into new (to me) techniques and ideas. I always learn a lot. In our blogging project, one person takes lead and writes a short summary of the paper and everyone else very briefly answers these questions:
- What’s your takeaway from the paper?
- What’s the coolest thing about it?
- What questions are you left with?
Joane started off the series in January and I followed this month talking about one of her papers.
The Homebrew Series: Inferring Demographic History With ABC, By Joane Elleouet And Sally Aitken
Want to know about the history of the populations you’re studying? Joane Elleouet and Sally Aitken see how far Approximate Bayesian Computation (ABC) and your sequencing method of choice can take you in a new paper in Molecular Ecology Resources. [more]
A long migration
5000 years ago, Native Americans in what is now the southern US created an extraordinary earthwork: Watson Brake took centuries to build and is the oldest mound complex we know about in North America.
We don’t know what it was called or what it meant to its builders; things that happened 5000 years ago are hard to reconstruct. But we try anyway.
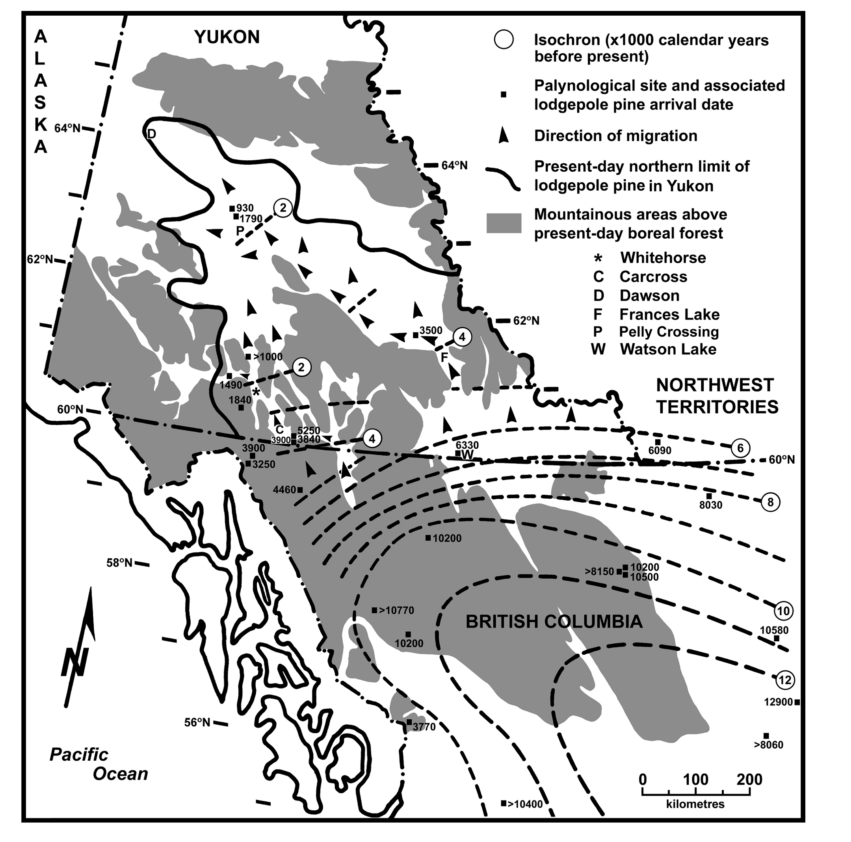
While the creators of Watson Brake were mixing their fish bones into the mounds in Louisiana, the eastern edge of the lodgepole pine migration front was crossing the Yukon Plateau 4000 km away. By the time the last handfuls of soil were being packed onto Watson Brake’s 25 foot tall ridges, lodgepole had hit the high, cold teeth of the Selwyn and Pelly Mountain ranges.

Photo by Ed Struzik of Cirque of the Unclimbables (actually in the Mackenzie Mountains to the east of the Selwyns)
The trees couldn’t cross the high mountains, but they followed the Frances River through a pass just 20 km wide to Frances Lake in southeastern Yukon.
4000 years ago the vizier to Pharaoh Mentuhotep IV led an expedition to Wadi Hammamat where he gave offerings to Min, a fertility god. A few years later, Mentuhotep was dead, childless, and the vizier was Pharaoh. He probably didn’t kill Mentuhotep, but that was 4000 years ago, so who knows?
About the same time Amenemhat was maybe-but-probably-not thinking about usurping the throne of ancient Egypt, lodgepole pushed out of Frances Lake’s narrow valley. Freed from the confines of the high mountains, lodgepole began a slow march up the the Tintina Trench.
The Tintina trench and its extension, the Northern Rocky Mountain Trench, are a longstanding human travel route and geological marvel. I wonder if the trees followed the people or the people followed the trees?
When the first Roman Emperor died, the eastern edge of the lodgepole migration front rounded the top edge of the Pelly Mountains at Pelly Crossing and sprawled across the eastern portions of the Yukon River watershed, down to meet the western migration front slowly making its way through the valleys of the northern Coast Range.

Pelly Crossing. Bridge not available for use by trees.
But lodgepole kept going up the Tintina Trench, too. Today, they’re are all the way up to Dawson. It’s not clear exactly when they got there, but the Tr’ondëk Hwëch’in have definitely been there longer than lodgepole has.
People move so much faster than trees that we don’t always notice their incredible journeys. But journey they do!
If you want the details and a discussion of uncertainty around these estimates, check out the paper: Strong, W. L., & Hills, L. V. (2013). Holocene migration of lodgepole pine (Pinus contorta var. latifolia) in southern Yukon, Canada. The Holocene, 23(9), 1340–1349. http://doi.org/10.1177/0959683613484614
Problems setting root.dir in knitr
RStudio sets the working directory to the project directory, but knitr sets the working directory to the .Rmd file directory. This creates issues when you are sourcing files relative to the project directory in your R markdown file. Specifically, knitr tells you it can’t find those files:
Error in file(filename, "r", encoding = encoding) : cannot open the connection In addition: Warning message: In file(filename, "r", encoding = encoding) : cannot open file 'home/sus/Documents/research_phd/analysis/phenodata_explore.R': No such file or directory
knitr has an option for dealing with this – root.dir – and Phil Mike Jones even gives a nice little knitr chunk example of using it.
But when I tried it, knitr kept failing to find my file.
```{r "setup", include=FALSE}
require("knitr")
opts_knit$set(root.dir = "~/Documents/research_phd/")
source('analysis/someanalysis.R')
```
It turns out I should have read the documentation more closely.
Knitr’s settings must be set in a chunk before any chunks which rely on those settings to be active. It is recommended to create a knit configuration chunk as the first chunk in a script with
cache = FALSEandinclude = FALSEoptions set. This chunk must not contain any commands which expect the settings in the configuration chunk to be in effect at the time of execution.
Breaking it into two chunks separating the knitr configuration from the sourcing solved the problem.
```{r "knitr config", cache = FALSE, include=FALSE}
require("knitr")
opts_knit$set(root.dir = "~/Documents/research_phd/")
```
```{r, "setup", echo = FALSE}
source('analysis/someanalysis.R')
```