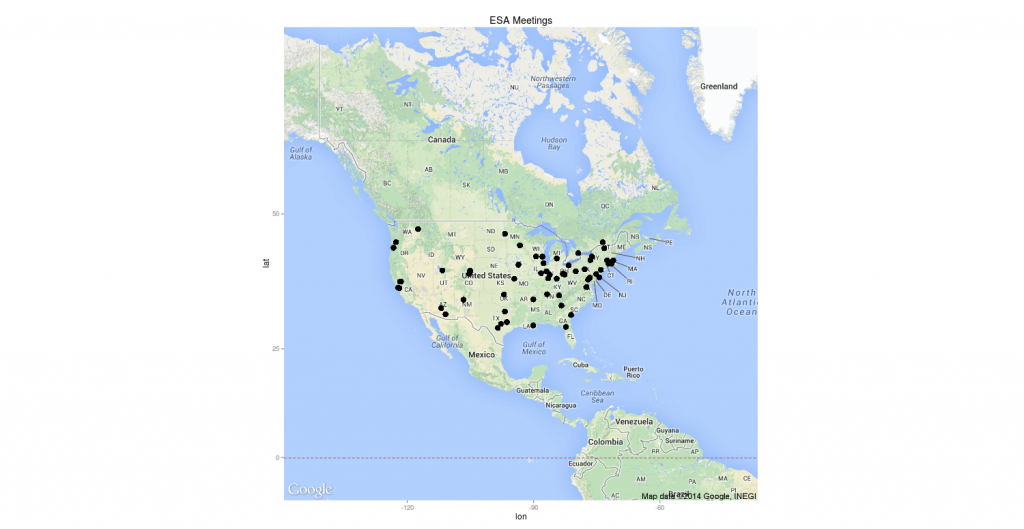
Locations of all Annual Meetings of the Ecological Society of America
In my spare time, I help ESA’s Historical Records Committee out with their website. I thought it would be nice to have a map of all of ESA’s annual meetings on the site. I downloaded the list of ESA Annual Meeting locations from the HRC website and got to work in R.
The biggest hurdle for me was translating the names of locations to latitude and longitude to add points to my ggmap. Luckily, someone else had written some functions that really helped with this.
getDocNodeVal=function(doc, path)
{
sapply(getNodeSet(doc, path), function(el) xmlValue(el))
}
gGeoCode=function(str)
{
library(XML)
u=paste('http://maps.google.com/maps/api/geocode/xml?sensor=false&address=',str)
doc = xmlTreeParse(u, useInternal=TRUE)
str=gsub(' ','%20',str)
lng=getDocNodeVal(doc, "/GeocodeResponse/result/geometry/location/lat")
lat=getDocNodeVal(doc, "/GeocodeResponse/result/geometry/location/lng")
c(lat,lng)
}
This code has a tiny error in it that caused me a bit of confusion – latitude is set to longitude and vice versa – but other than that it works great at taking a city name and churning out coordinates. That is, unless you have commas in your string or try to submit a bunch at once.
If the string you submit to gGeoCode has commas in it, it will return list() instead of coordinates.
If you try to use gGeoCode with a whole bunch of cities using a for loop or apply function it will return NULL for some or all cities because you’re asking Google for too many results too quickly (I think). I got around that by using Sys.sleep in a for loop.
getlatlong <- function(locations) { #get lat and long for each location using gGeoCode
dat <- cbind(lat=numeric(0), long=numeric(0))
for (i in 1:length(locations)) {
latlon <- gGeoCode(locations[i])[c(1:2)] #choose only the first location returned.
dat <- rbind(dat, latlon, deparse.level=0)
Sys.sleep(.5) #returns null values without this
}
dat <- data.frame(location=locations, dat)
}
You can get the complete code in the github repo here.
